This article describes how to compute two samples Wilcoxon test in R.
Visualize your data and compute Wilcoxon test in R
R function to compute Wilcoxon test
To perform two-samples Wilcoxon test comparing the means of two independent samples (x & y), the R function wilcox.test() can be used as follow:
wilcox.test(x, y, alternative = "two.sided")- x,y: numeric vectors
- alternative: the alternative hypothesis. Allowed value is one of “two.sided” (default), “greater” or “less”.
Import your data into R
Prepare your data as specified here: Best practices for preparing your data set for R
Save your data in an external .txt tab or .csv files
Import your data into R as follow:
# If .txt tab file, use this
my_data <- read.delim(file.choose())
# Or, if .csv file, use this
my_data <- read.csv(file.choose())Here, we’ll use an example data set, which contains the weight of 18 individuals (9 women and 9 men):
# Data in two numeric vectors
women_weight <- c(38.9, 61.2, 73.3, 21.8, 63.4, 64.6, 48.4, 48.8, 48.5)
men_weight <- c(67.8, 60, 63.4, 76, 89.4, 73.3, 67.3, 61.3, 62.4)
# Create a data frame
my_data <- data.frame(
group = rep(c("Woman", "Man"), each = 9),
weight = c(women_weight, men_weight)
)We want to know, if the median women’s weight differs from the median men’s weight?
Check your data
print(my_data) group weight
1 Woman 38.9
2 Woman 61.2
3 Woman 73.3
4 Woman 21.8
5 Woman 63.4
6 Woman 64.6
7 Woman 48.4
8 Woman 48.8
9 Woman 48.5
10 Man 67.8
11 Man 60.0
12 Man 63.4
13 Man 76.0
14 Man 89.4
15 Man 73.3
16 Man 67.3
17 Man 61.3
18 Man 62.4It’s possible to compute summary statistics (median and interquartile range (IQR)) by groups. The dplyr package can be used.
- To install dplyr package, type this:
install.packages("dplyr")- Compute summary statistics by groups:
library(dplyr)
group_by(my_data, group) %>%
summarise(
count = n(),
median = median(weight, na.rm = TRUE),
IQR = IQR(weight, na.rm = TRUE)
)Source: local data frame [2 x 4]
group count median IQR
(fctr) (int) (dbl) (dbl)
1 Man 9 67.3 10.9
2 Woman 9 48.8 15.0Visualize your data using box plots
You can draw R base graphs as described at this link: R base graphs. Here, we’ll use the ggpubr R package for an easy ggplot2-based data visualization
- Install the latest version of ggpubr from GitHub as follow (recommended):
# Install
if(!require(devtools)) install.packages("devtools")
devtools::install_github("kassambara/ggpubr")- Or, install from CRAN as follow:
install.packages("ggpubr")- Visualize your data:
# Plot weight by group and color by group
library("ggpubr")
ggboxplot(my_data, x = "group", y = "weight",
color = "group", palette = c("#00AFBB", "#E7B800"),
ylab = "Weight", xlab = "Groups")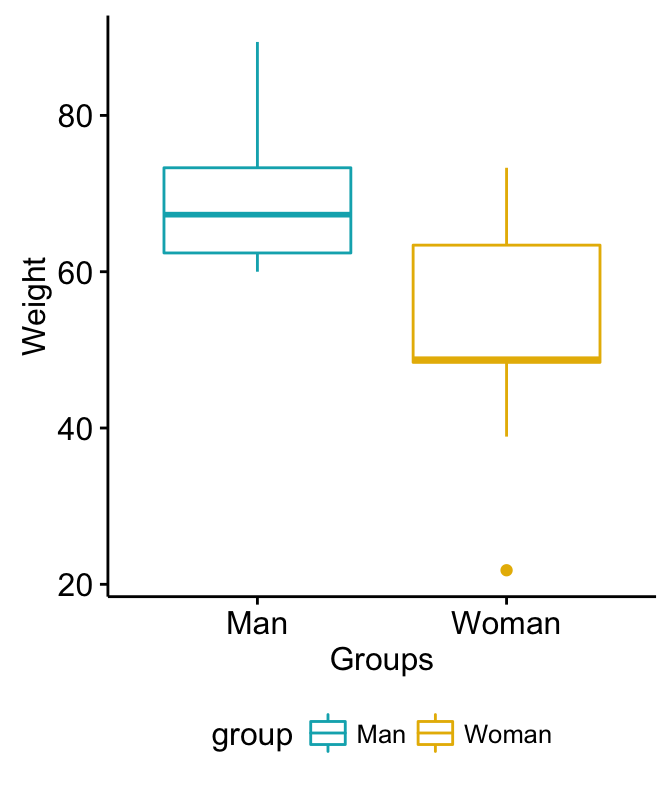
Unpaired Two-Samples Wilcoxon Test in R
Compute unpaired two-samples Wilcoxon test
Question : Is there any significant difference between women and men weights?
1) Compute two-samples Wilcoxon test - Method 1: The data are saved in two different numeric vectors.
res <- wilcox.test(women_weight, men_weight)
res
Wilcoxon rank sum test with continuity correction
data: women_weight and men_weight
W = 15, p-value = 0.02712
alternative hypothesis: true location shift is not equal to 0It will give a warning message, saying that “cannot compute exact p-value with tie”. It comes from the assumption of a Wilcoxon test that the responses are continuous. You can suppress this message by adding another argument exact = FALSE, but the result will be the same.
2) Compute two-samples Wilcoxon test - Method 2: The data are saved in a data frame.
res <- wilcox.test(weight ~ group, data = my_data,
exact = FALSE)
res
Wilcoxon rank sum test with continuity correction
data: weight by group
W = 66, p-value = 0.02712
alternative hypothesis: true location shift is not equal to 0# Print the p-value only
res$p.value[1] 0.02711657As you can see, the two methods give the same results.
The p-value of the test is 0.02712, which is less than the significance level alpha = 0.05. We can conclude that men’s median weight is significantly different from women’s median weight with a p-value = 0.02712.
Note that:
- if you want to test whether the median men’s weight is less than the median women’s weight, type this:
wilcox.test(weight ~ group, data = my_data,
exact = FALSE, alternative = "less")- Or, if you want to test whether the median men’s weight is greater than the median women’s weight, type this
wilcox.test(weight ~ group, data = my_data,
exact = FALSE, alternative = "greater")Online unpaired two-samples Wilcoxon test calculator
You can perform unpaired two-samples Wilcoxon test, online, without any installation by clicking the following link:
See also
- Compare one-sample mean to a standard known mean
- Compare the means of two independent groups
Infos
This analysis has been performed using R software (ver. 3.2.4).

















