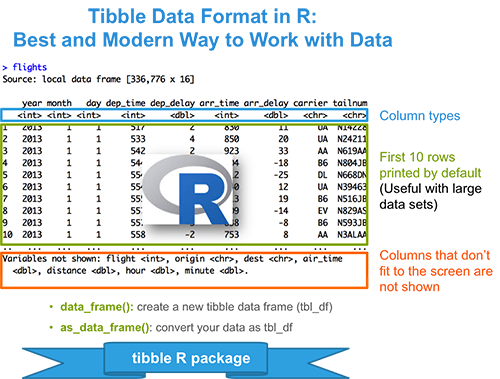
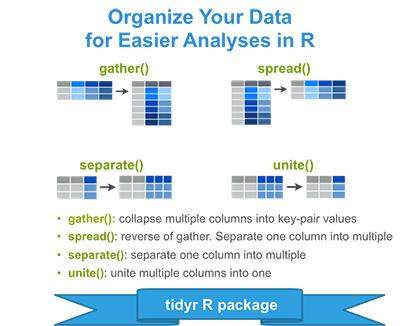
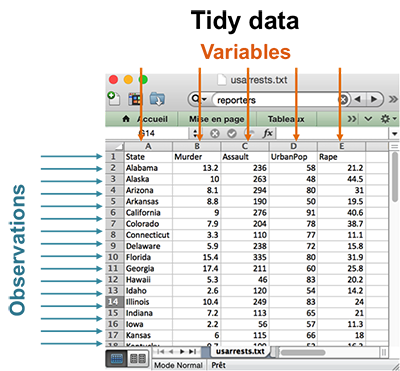
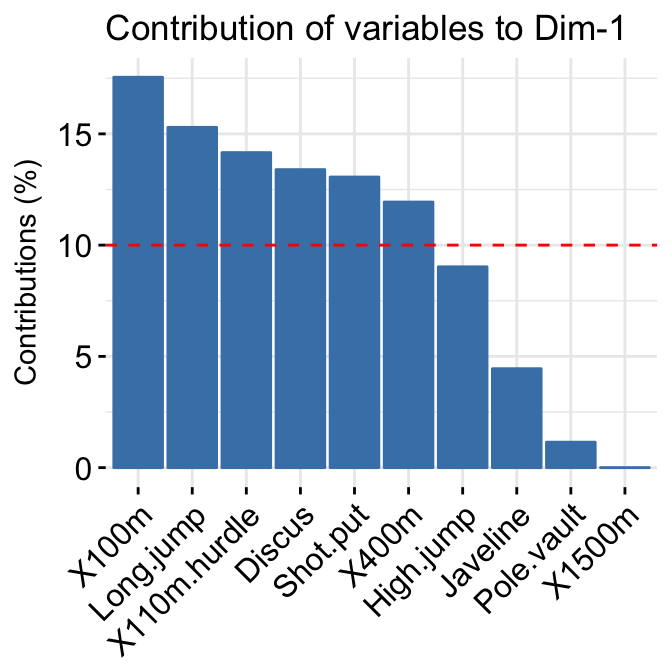
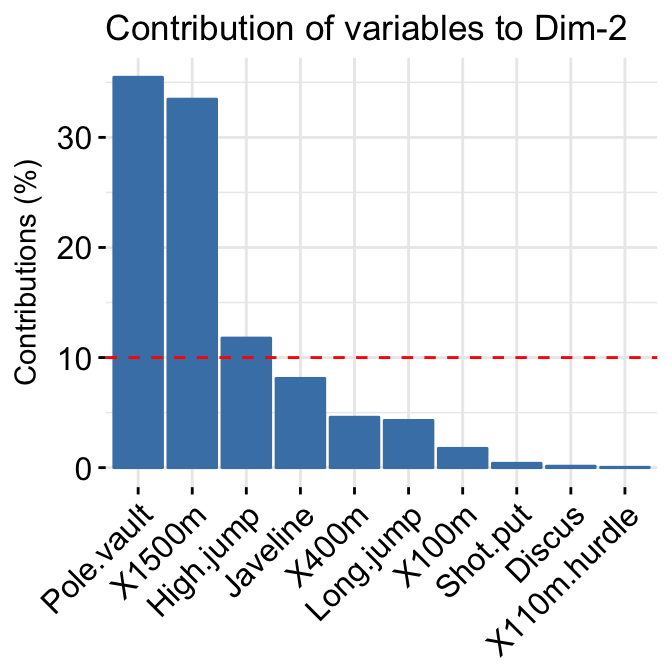
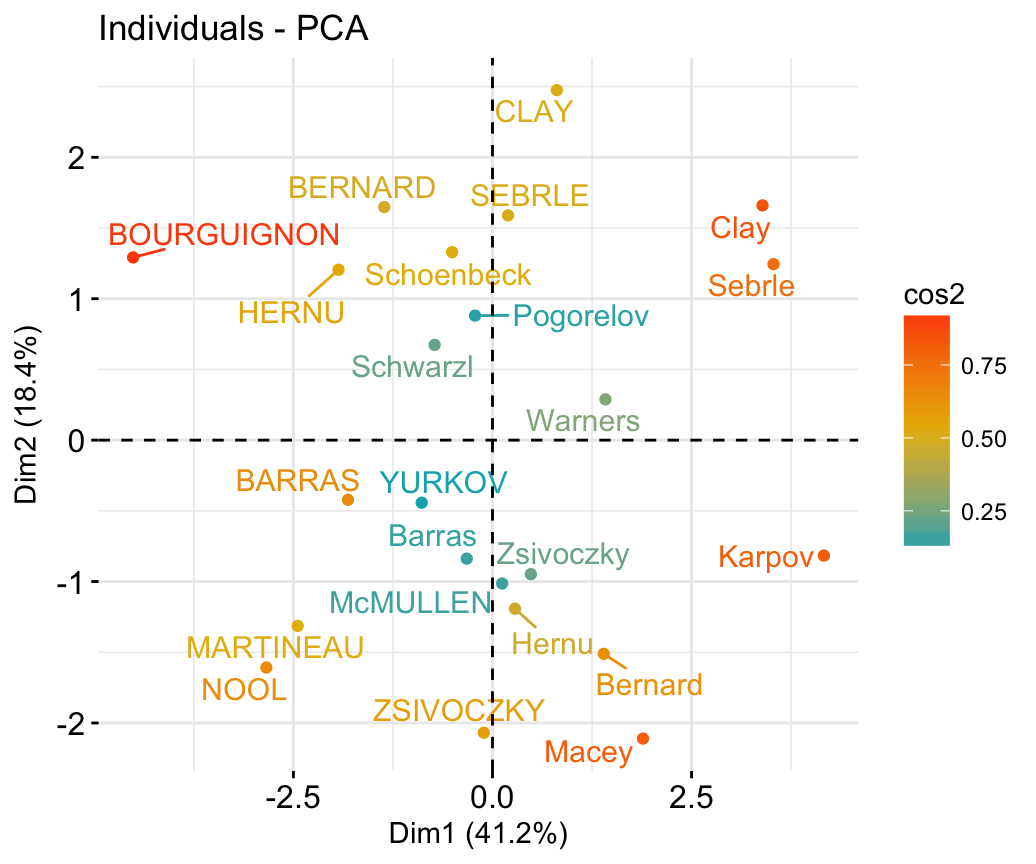
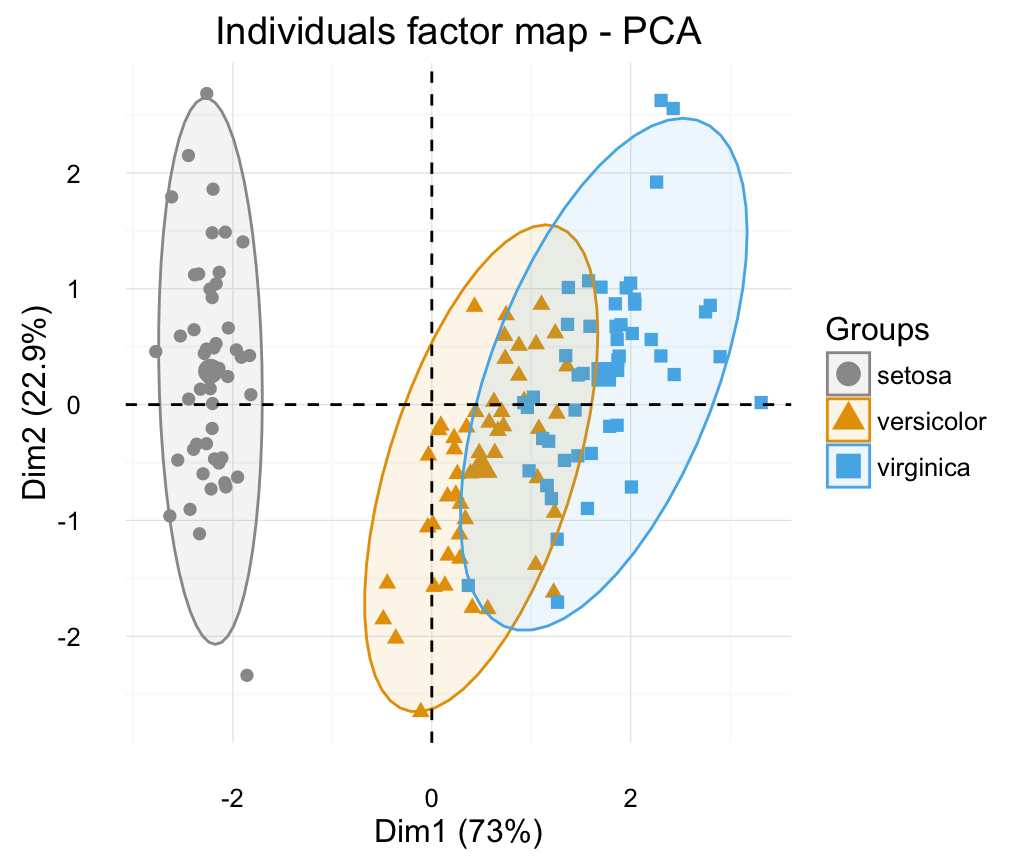
In the previous chapters we described the essentials of R programming as well as how to import data into R. Here, youll learn how to export data from R to txt, csv, Excel (xls, xlsx) and R data file formats. Additionally, well describe how to create and format Word and PowerPoint documents from R.
- Export Data From R to txt|csv|Excel files
- Create and format word and powerpoint documents using R and ReporteRs package:
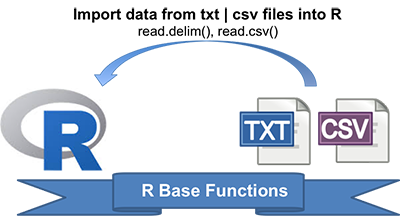
- R base functions for writing data: write.table(), write.csv(), write.csv2()
- Writing data to a file
# Loading mtcars data
data("mtcars")
# Write data to txt file: tab separated values
# sep = "\t"
write.table(mtcars, file = "mtcars.txt", sep = "\t",
row.names = TRUE, col.names = NA)
# Write data to csv files:
# decimal point = "." and value separators = comma (",")
write.csv(mtcars, file = "mtcars.csv")
# Write data to csv files:
# decimal point = comma (",") and value separators = semicolon (";")
write.csv2(mtcars, file = "mtcars.csv")Read more: Writing data from R to a txt|csv file: R base functions
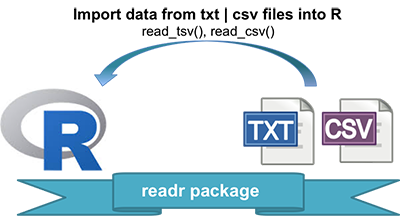
- Installing and loading readr: install.packages(readr)
- readr functions for writing data: write_tsv(), write_csv()
- Writing data to a file
# Loading mtcars data
data("mtcars")
library("readr")
# Writing mtcars data to a tsv file
write_tsv(mtcars, path = "mtcars.txt")
# Writing mtcars data to a csv file
write_csv(mtcars, path = "mtcars.csv")Read more: Fast writing of Data From R to txt|csv Files: readr package
- Installing xlsx package: install.packages(xlsx)
- Using xlsx package: write.xlsx()
library("xlsx")
# Write the first data set in a new workbook
write.xlsx(USArrests, file = "myworkbook.xlsx",
sheetName = "USA-ARRESTS", append = FALSE)
# Add a second data set in a new worksheet
write.xlsx(mtcars, file = "myworkbook.xlsx",
sheetName="MTCARS", append=TRUE)Read more: Writing data from R to Excel files (xls|xlsx)
- Save one object to a file: saveRDS(object, file), readRDS(file)
- Save multiple objects to a file: save(data1, data2, file), load(file)
- Save your entire workspace: save.image(), load()
- Saving and restoring one single R object:
# Save a single object to a file
saveRDS(mtcars, "mtcars.rds")
# Restore it under a different name
my_data <- readRDS("mtcars.rds")- Saving and restoring one or more R objects:
# Save multiple objects
save(data1, data2, file = "data.RData")
# To load the data again
load("data.RData")- Saving and restoring your entire workspace:
# Save your workspace
save.image(file = "my_work_space.RData")
# Load the workspace again
load("my_work_space.RData")Read more: Saving data into R data format: RDATA and RDS
ReporteRs package, by David Gohel, provides easy to use functions to write and formatWord documents. It can be also used to generate Word document from a template file with logos, fonts, etc. ReporteRs is Java-based solution, so it works on Windows, Linux and Mac OS systems.
- Install and load the ReporteRs R package
- Create a simple Word document
- Add texts : title and paragraphs of texts
- Format the text of a Word document using R software
- Add plots and images
- Add a table
- Add lists : ordered and unordered lists
- Add a footnote to a Word document
- Add R scripts
- Add a table of contents into a Word document
Read more: Create and format Word documents with R and ReporteRs package
- Quick introduction to ReporteRs package
- Create a Word document using a template file

Read more: Create a Word document from a template file with R and ReporteRs package
- Add a simple table
- Add a formatted table
- Change the background colors of rows and columns
- Change cell background and text colors
- Insert content into a table : header and footer rows
- Analyze, format and export a correlation matrix into a Word document
- Powerpoint
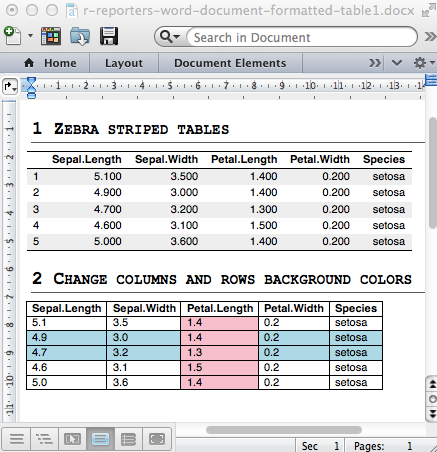
Read more: Add a table into a Word document with R and ReporteRs package
- Why is it important to be able to generate a PowerPoint report from R ?
- Reason I : Many collaborators works with Microsoft office tools
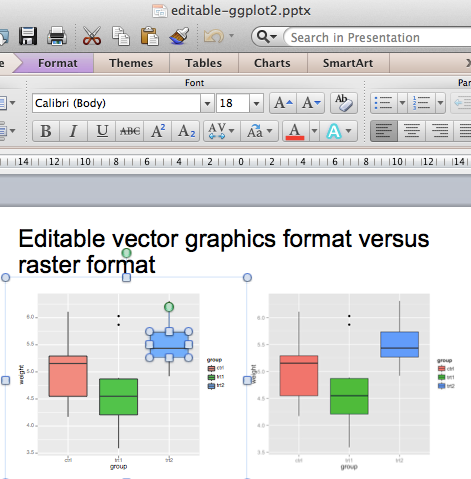
- Reason II : keeping beautiful R graphs beautiful for publications
- Install and load the ReporteRs package
- Create a simple PowerPoint document
- Slide layout
- Generate a simple PowerPoint document from R software
- Format the text of a PowerPoint document
- Add plots and images
- Add a table
- Add ordered and unordered lists
- Create a PowerPoint document from a template file

Read more: Create and format PowerPoint documents with R and ReporteRs
- Case of base graphs
- Case of graphs generated using ggplot2