Survival analysis focuses on the expected duration of time until occurrence of an event of interest. However, this failure time may not be observed within the study time period, producing the so-called censored observations.
The R package survival fits and plots survival curves using R base graphs. There are also several R packages/functions for drawing survival curves using ggplot2 system:
These packages/functions are limited:
The default graph generated with the R package survival is ugly and it requires programming skills for drawing a nice looking survival curves. There is no option for displaying the number at risk table.
GGally and ggfortify dont contain any option for drawing the number at risk table. You need also some knowledge in ggplot2 plotting system for drawing a ready-to-publish survival curves.
- There are different version of the function ggkm() on the web. Most of them are not updated and dont work with the current version of ggplot2.
Here, we developed and present the survminer R package for facilitating survival analysis and visualization.
survminer - Main features
The current version contains the function ggsurvplot() for easily drawing beautiful and ready-to-publish survival curves using ggplot2. ggsurvplot() includes also some options for displaying the p-value and the number at risk table, under the survival curves.
Installation and loading
Install from CRAN:
install.packages("survminer")Or, install the latest version from GitHub:
# Install
if(!require(devtools)) install.packages("devtools")
devtools::install_github("kassambara/survminer")# Loading
library("survminer")Getting started
The R package survival is required for fitting survival curves.
Draw survival curves without grouping
# Fit survival curves
require("survival")
fit <- survfit(Surv(time, status) ~ 1, data = lung)
# Drawing curves
ggsurvplot(fit, color = "#2E9FDF")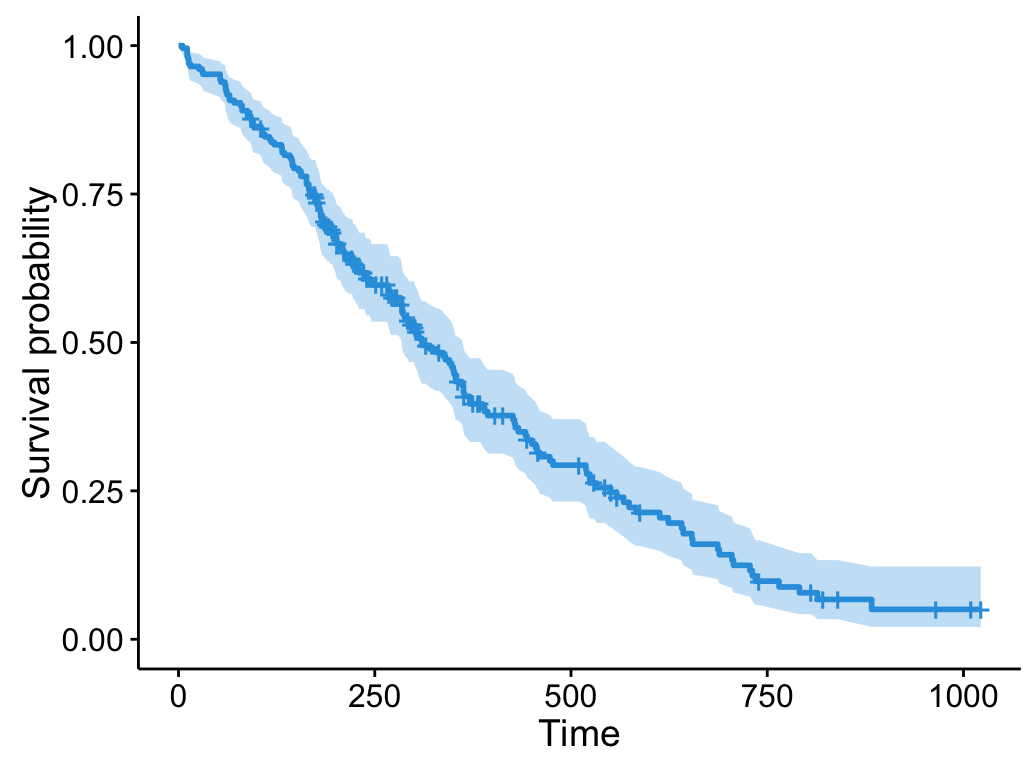
Draw survival curves with two groups
Basic plots
# Fit survival curves
require("survival")
fit<- survfit(Surv(time, status) ~ sex, data = lung)
# Drawing survival curves
ggsurvplot(fit)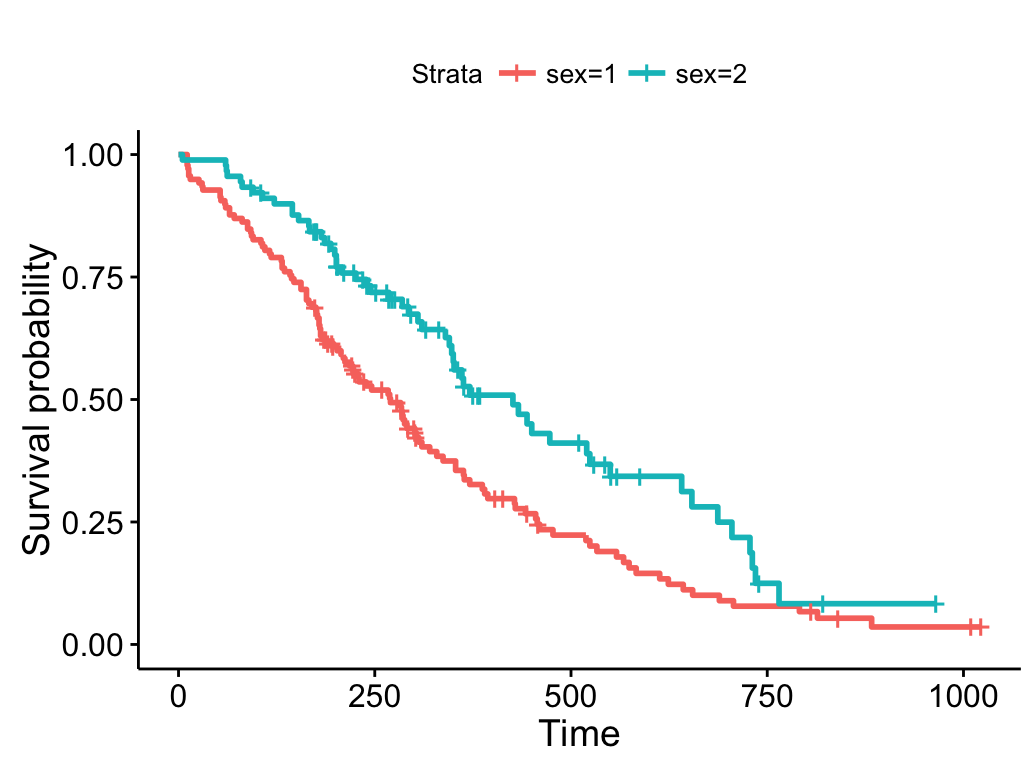
Change legend title, labels and position
# Change the legend title and labels
ggsurvplot(fit, legend = "bottom",
legend.title = "Sex",
legend.labs = c("Male", "Female"))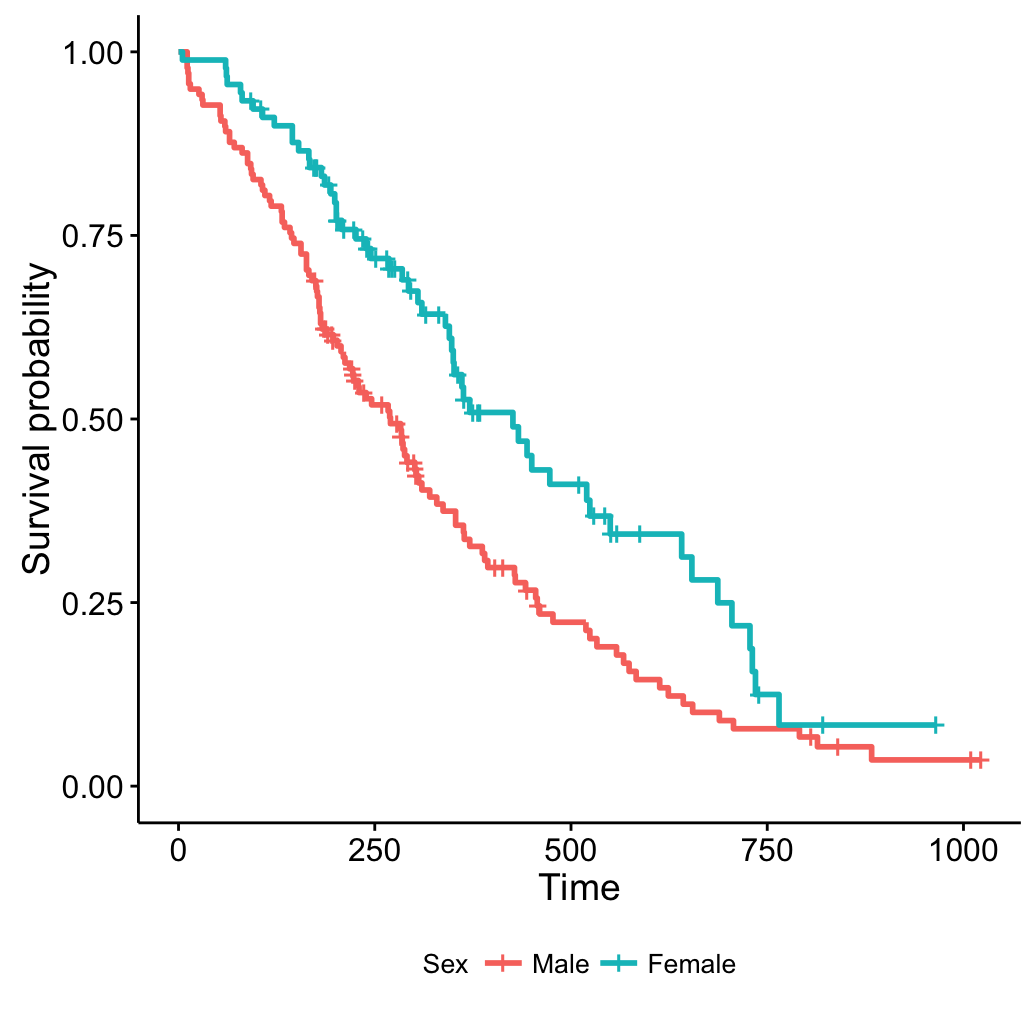
# Specify legend position by its coordinates
ggsurvplot(fit, legend = c(0.2, 0.2))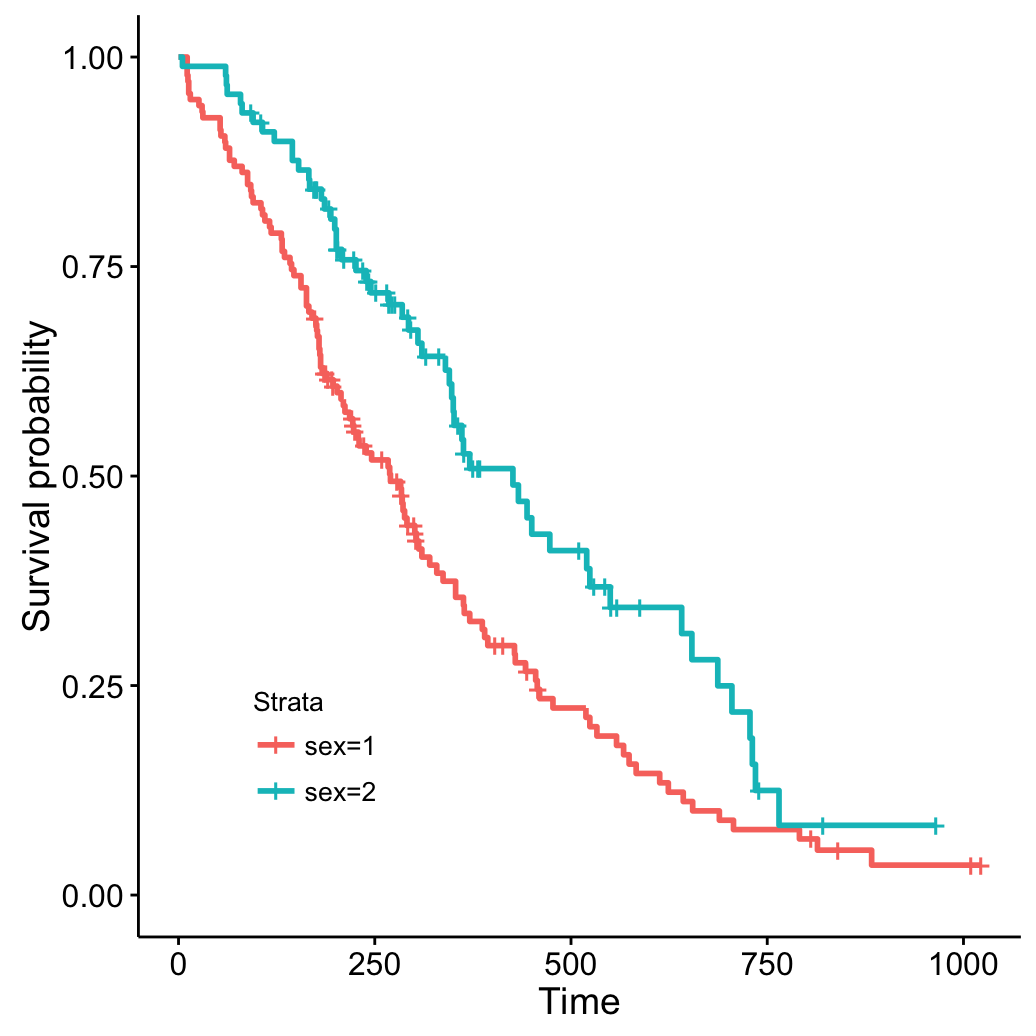
Change line types and color palettes
# change line size --> 1
# Change line types by groups (i.e. "strata")
# and change color palette
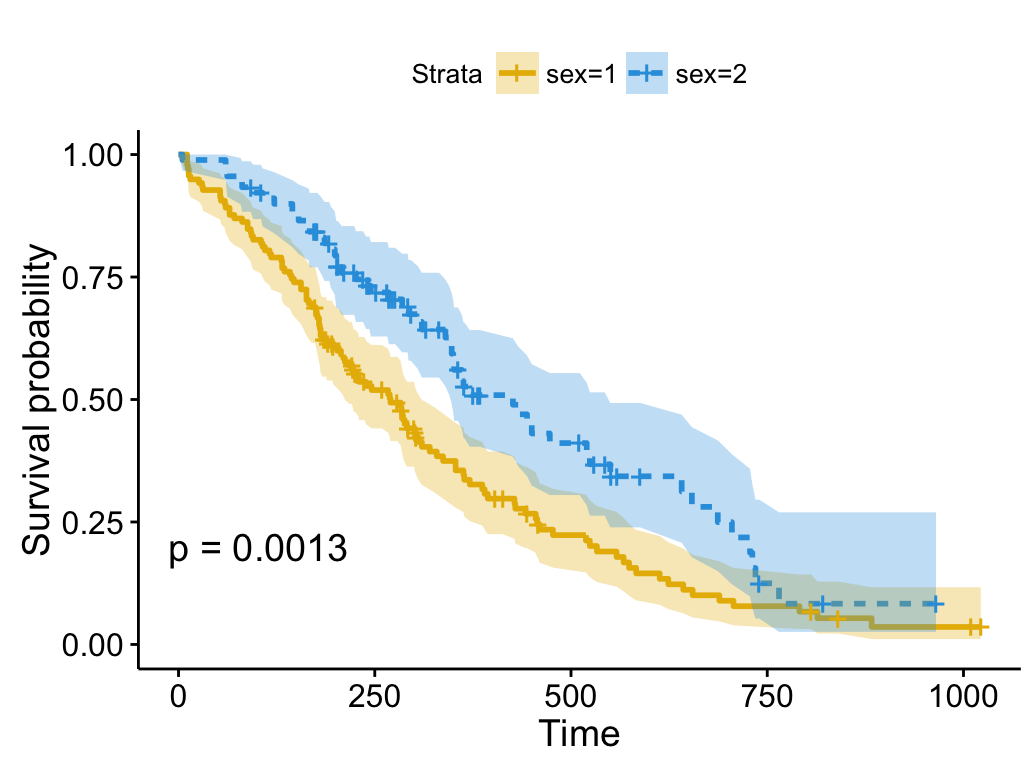
ggsurvplot(fit, size = 1, # change line size
linetype = "strata", # change line type by groups
break.time.by = 250, # break time axis by 250
palette = c("#E7B800", "#2E9FDF"), # custom color palette
conf.int = TRUE, # Add confidence interval
pval = TRUE # Add p-value
)
# Use brewer color palette "Dark2"
ggsurvplot(fit, linetype = "strata",
conf.int = TRUE, pval = TRUE,
palette = "Dark2")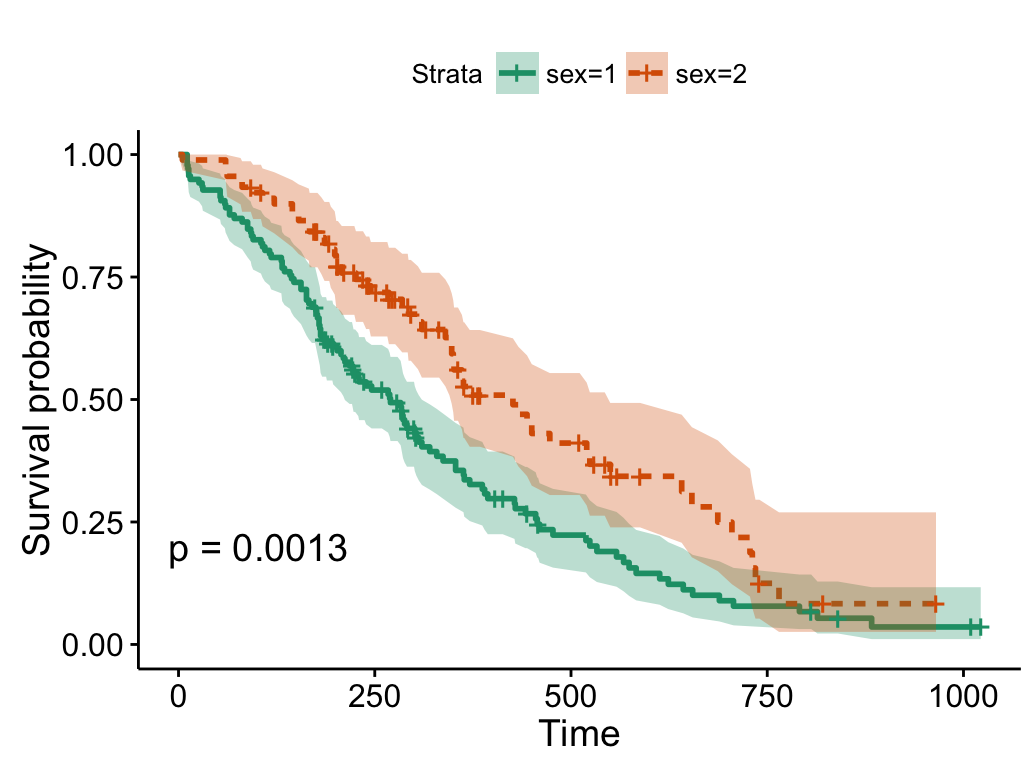
# Use grey palette
ggsurvplot(fit, linetype = "strata",
conf.int = TRUE, pval = TRUE,
palette = "grey")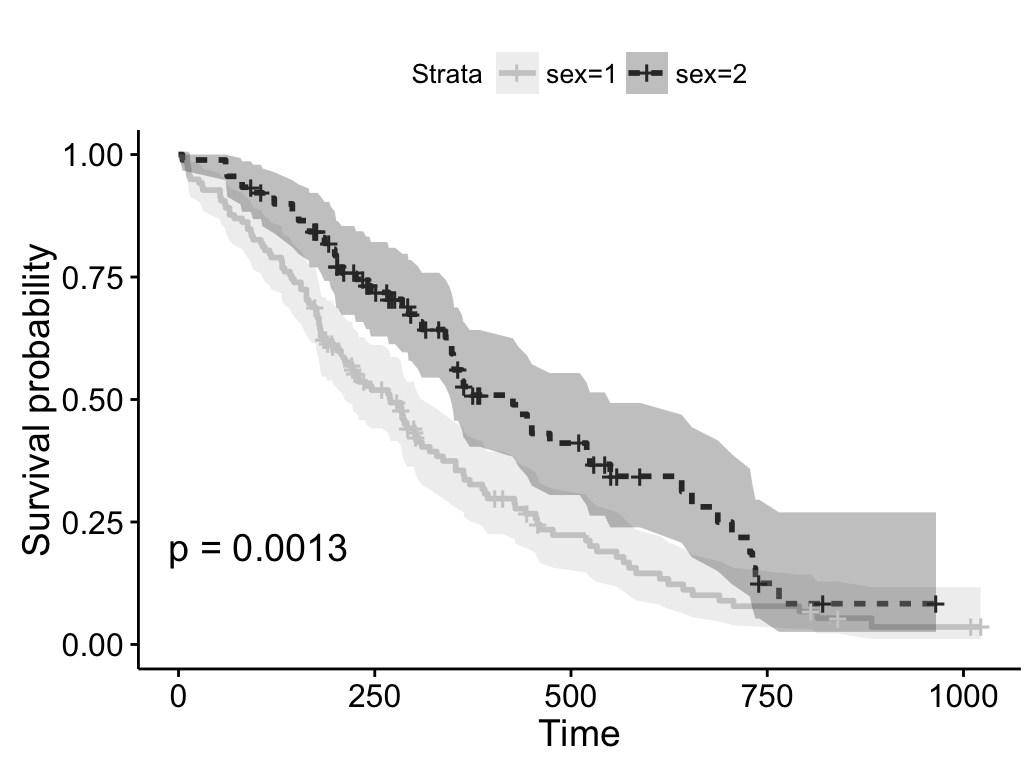
Add number at risk table
# Add Risk table
ggsurvplot(fit, pval = TRUE, conf.int = TRUE,
risk.table = TRUE)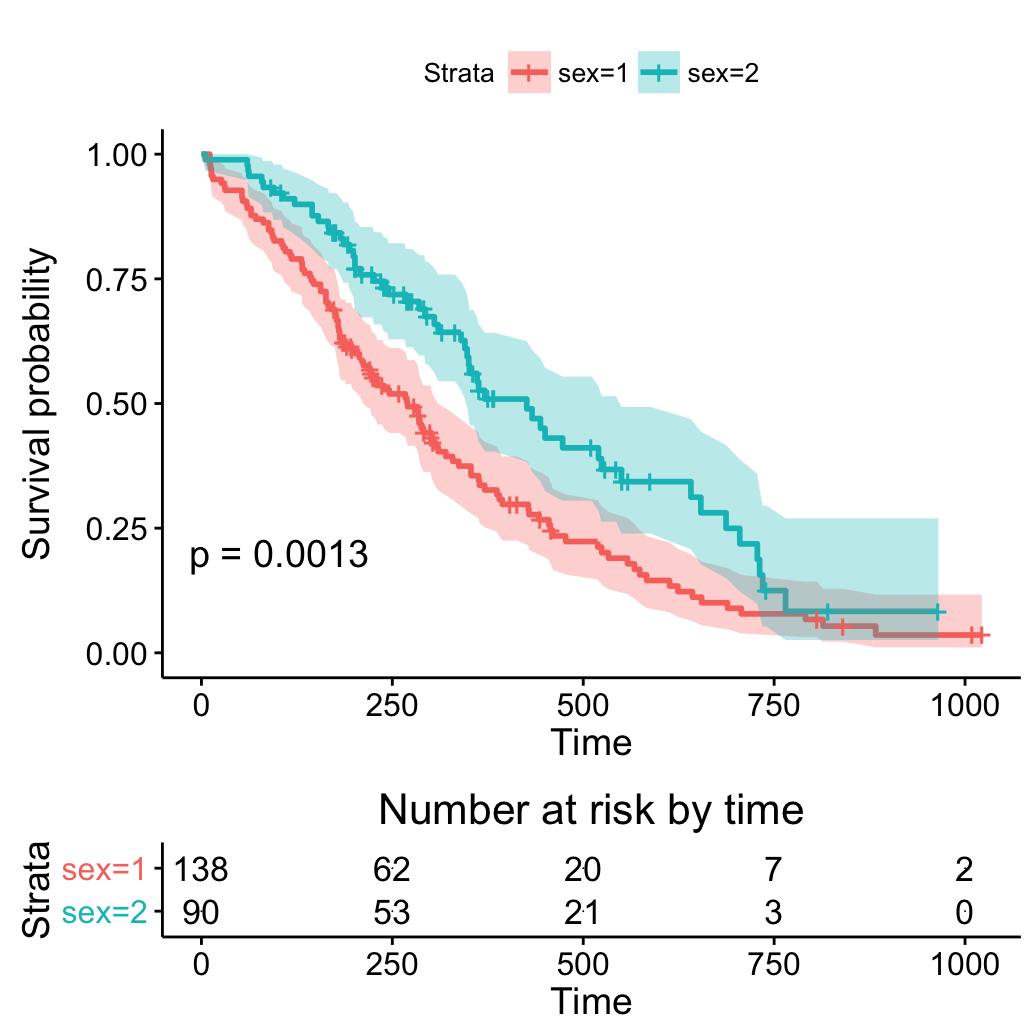
# Change color, linetype by strata, risk.table color by strata
ggsurvplot(fit,
pval = TRUE, conf.int = TRUE,
risk.table = TRUE, # Add risk table
risk.table.col = "strata", # Risk table color by groups
lienetype = "strata", # Change line type by groups
ggtheme = theme_bw(), # Change ggplot2 theme
palette = c("#E7B800", "#2E9FDF"))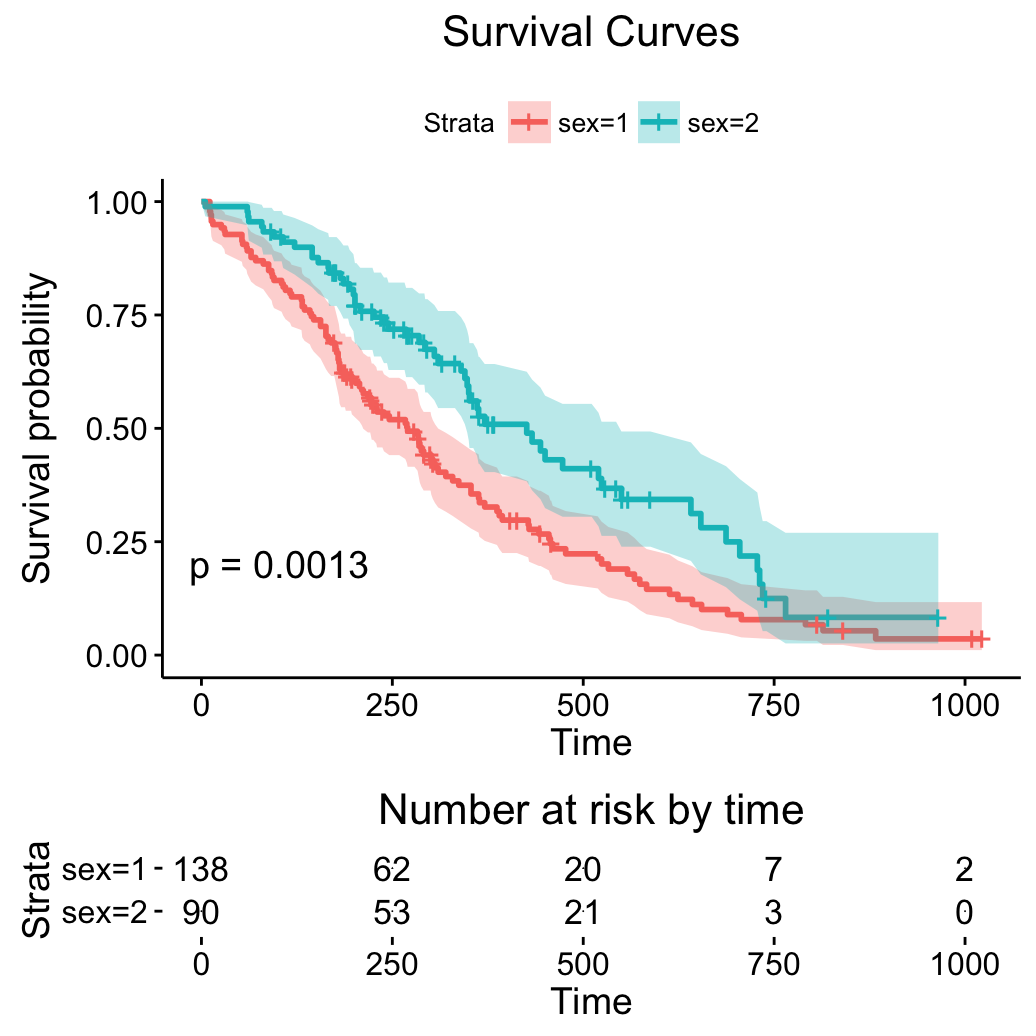
Transform survival curves: plot cumulative events and hazard function
# Plot cumulative events
ggsurvplot(fit, conf.int = TRUE,
palette = c("#FF9E29", "#86AA00"),
risk.table = TRUE, risk.table.col = "strata",
fun = "event")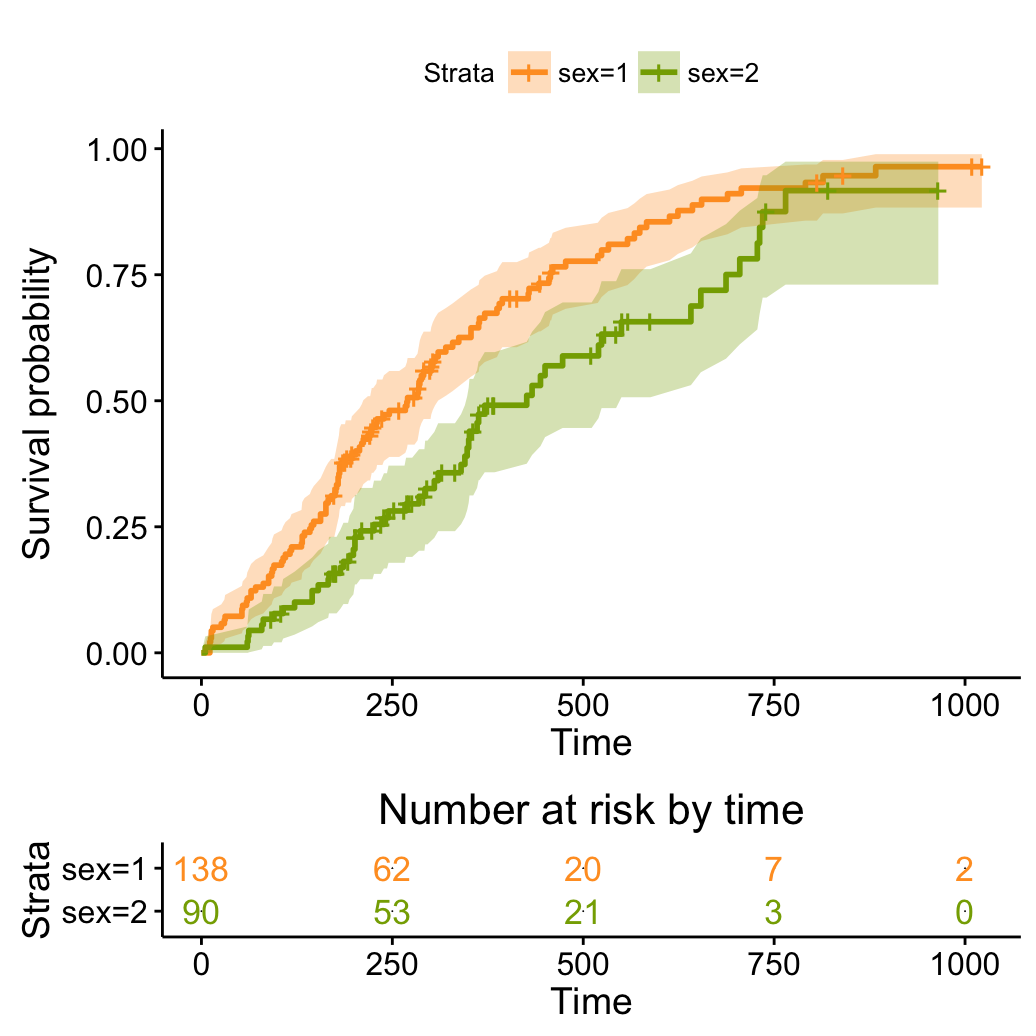
# Plot the cumulative hazard function
ggsurvplot(fit, conf.int = TRUE,
palette = c("#FF9E29", "#86AA00"),
risk.table = TRUE, risk.table.col = "strata",
fun = "cumhaz")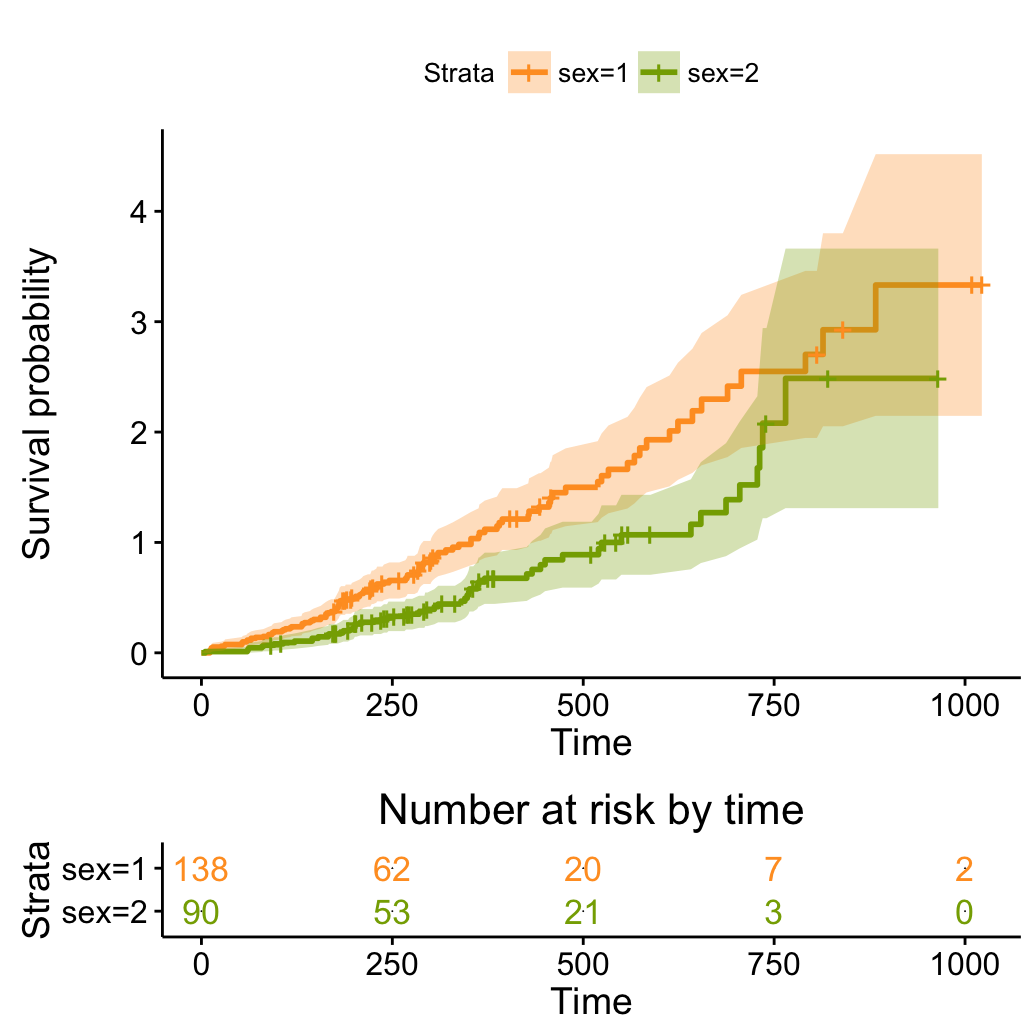
# Arbitrary function
ggsurvplot(fit, conf.int = TRUE,
palette = c("#FF9E29", "#86AA00"),
risk.table = TRUE, risk.table.col = "strata",
pval = TRUE,
fun = function(y) y*100)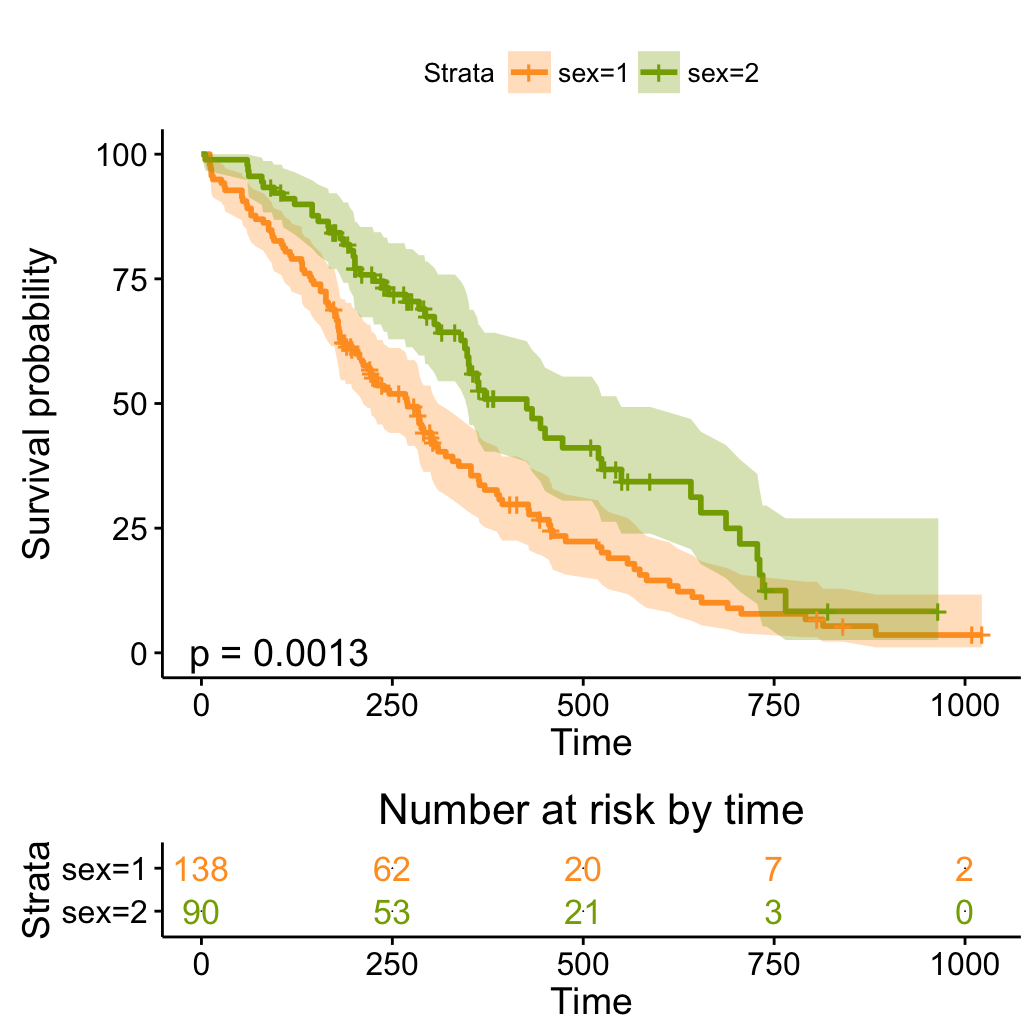
Survival curves with multiple groups
# Fit (complexe) survival curves
#++++++++++++++++++++++++++++++++++++
require("survival")
fit2 <- survfit( Surv(time, status) ~ rx + adhere,
data = colon )
# Visualize
#++++++++++++++++++++++++++++++++++++
# Visualize: add p-value, chang y limits
# change color using brewer palette
ggsurvplot(fit2, pval = TRUE,
break.time.by = 400,
risk.table = TRUE)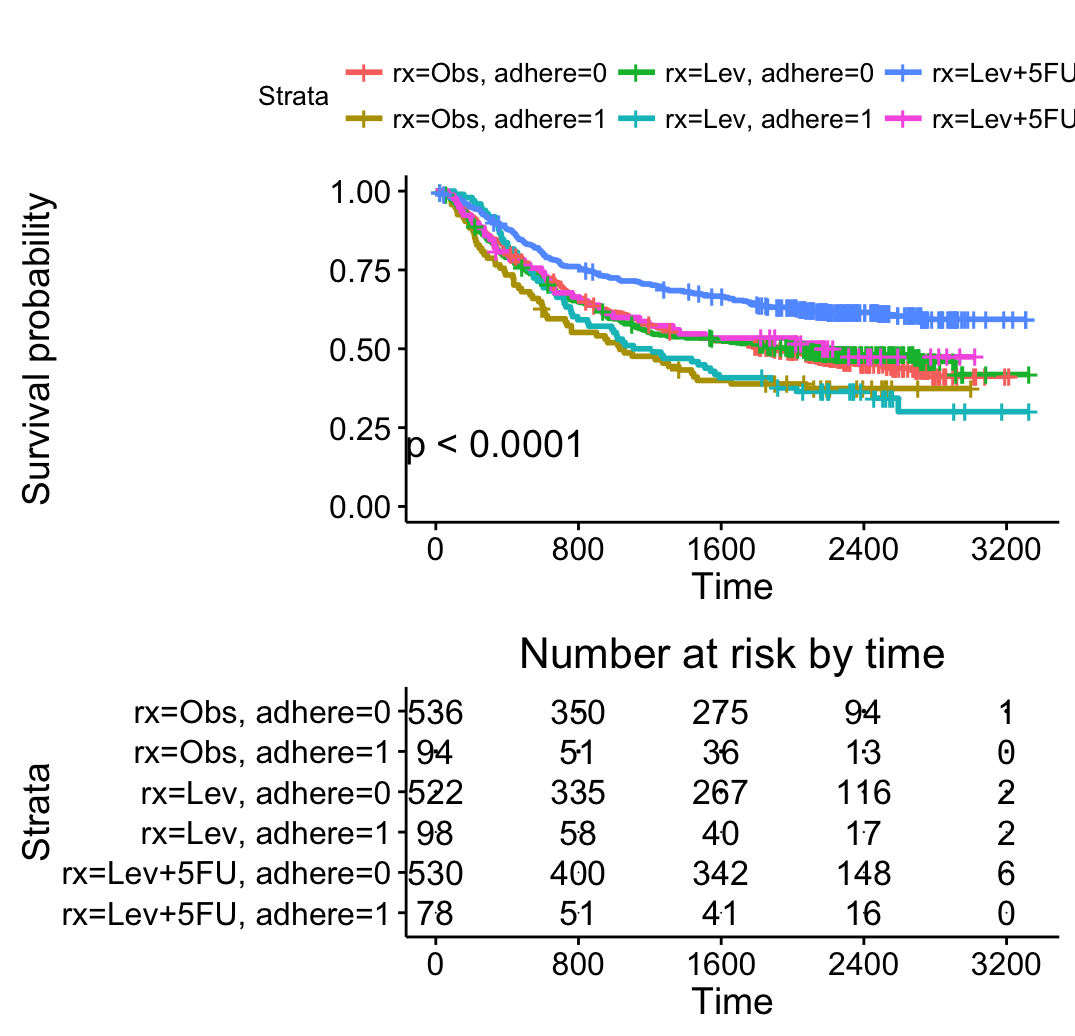
# Adjust risk table and survival plot locations
# ++++++++++++++++++++++++++++++++++++
# Adjust risk table location, shift to the left
ggsurvplot(fit2, pval = TRUE,
break.time.by = 400,
risk.table = TRUE,
risk.table.col = "strata",
risk.table.adj = -2, # risk table location adj
palette = "Dark2")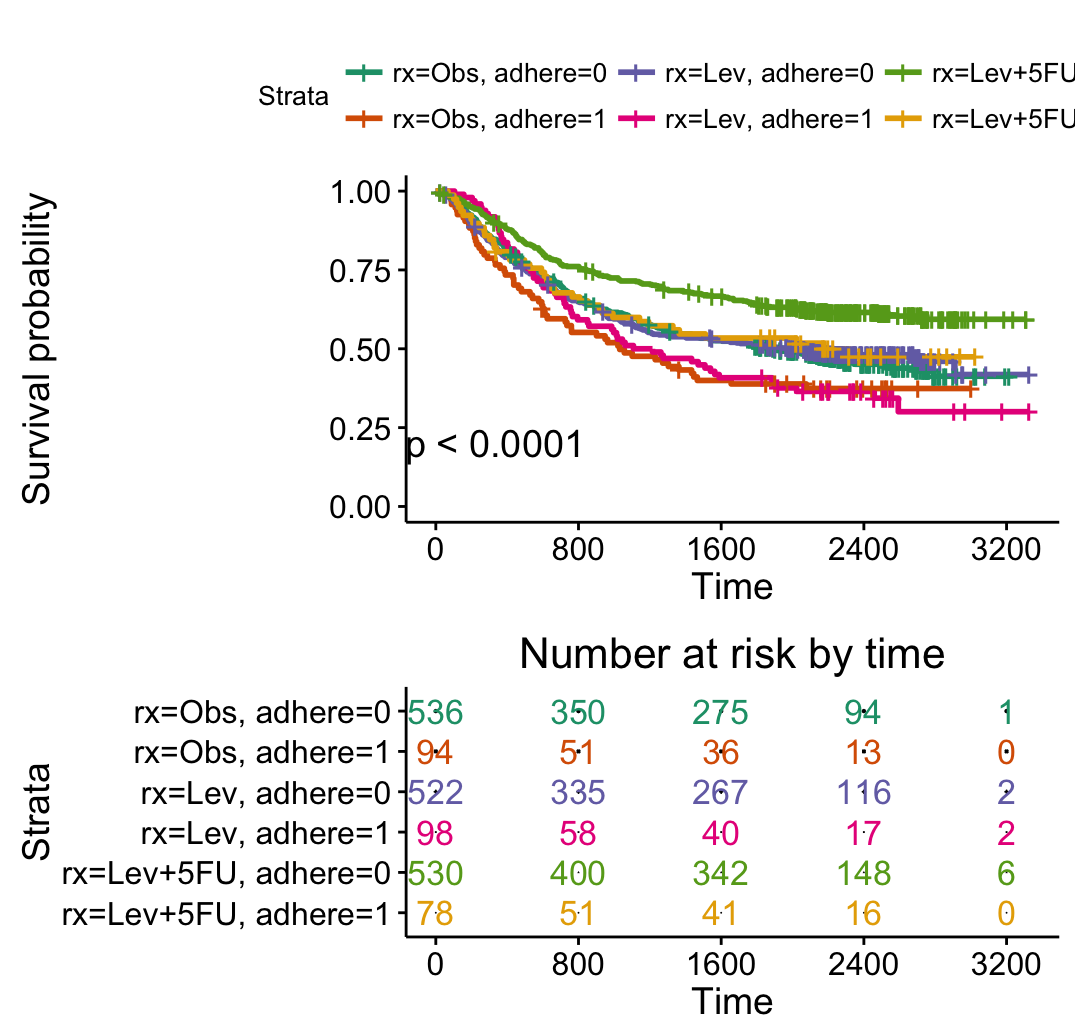
# Adjust survival plot location, shift to the right
# ++++++++++++++++++++++++++++++++++++
ggsurvplot(fit2, pval = TRUE,
break.time.by = 400,
risk.table = TRUE,
risk.table.col = "strata",
surv.plot.adj = 4.9, # surv plot location adj
palette = "Dark2")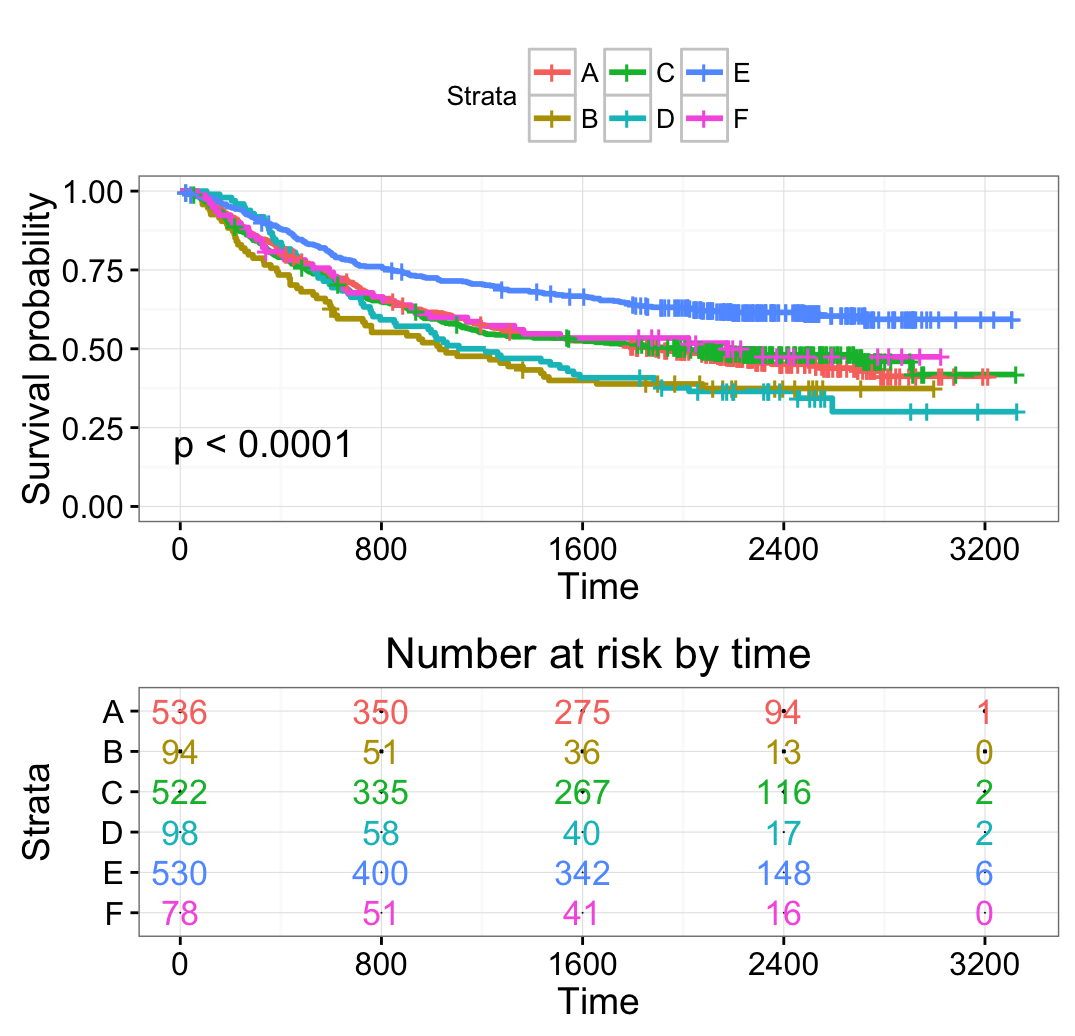
# Risk table height
# ++++++++++++++++++++++++++++++++++++
ggsurvplot(fit2, pval = TRUE,
break.time.by = 400,
risk.table = TRUE,
risk.table.col = "strata",
risk.table.height = 0.5, # Useful when you have multiple groups
surv.plot.adj = 4.9, # surv plot location adj
palette = "Dark2")
# Change legend labels
# ++++++++++++++++++++++++++++++++++++
ggsurvplot(fit2, pval = TRUE,
break.time.by = 400,
risk.table = TRUE,
risk.table.col = "strata",
ggtheme = theme_bw(),
legend.labs = c("A", "B", "C", "D", "E", "F"))
Infos
This article was built with:
## setting value
## version R version 3.2.3 (2015-12-10)
## system x86_64, darwin13.4.0
## ui X11
## language (EN)
## collate fr_FR.UTF-8
## tz Europe/Paris
## date 2016-01-17
##
## package * version date source
## colorspace 1.2-6 2015-03-11 CRAN (R 3.2.0)
## dichromat 2.0-0 2013-01-24 CRAN (R 3.2.0)
## digest 0.6.8 2014-12-31 CRAN (R 3.2.0)
## ggplot2 * 2.0.0 2015-12-18 CRAN (R 3.2.3)
## gridExtra 2.0.0 2015-07-14 CRAN (R 3.2.0)
## gtable 0.1.2 2012-12-05 CRAN (R 3.2.0)
## labeling 0.3 2014-08-23 CRAN (R 3.2.0)
## magrittr 1.5 2014-11-22 CRAN (R 3.2.0)
## MASS 7.3-45 2015-11-10 CRAN (R 3.2.3)
## munsell 0.4.2 2013-07-11 CRAN (R 3.2.0)
## plyr 1.8.3 2015-06-12 CRAN (R 3.2.0)
## RColorBrewer 1.1-2 2014-12-07 CRAN (R 3.2.0)
## Rcpp 0.12.2 2015-11-15 CRAN (R 3.2.2)
## reshape2 1.4.1 2014-12-06 CRAN (R 3.2.0)
## scales 0.3.0 2015-08-25 CRAN (R 3.2.0)
## stringi 1.0-1 2015-10-22 CRAN (R 3.2.0)
## stringr 1.0.0 2015-04-30 CRAN (R 3.2.0)
## survival * 2.38-3 2015-07-02 CRAN (R 3.2.3)
## survminer * 0.1.1 2016-01-17 local